Download Daymet#
Daymet provides gridded meteorological data for North American at 1km spatial resolution with daily timestep from 1980 ~ present. website and user guide
Available variables:
Variable |
Description (units) |
|---|---|
tmax |
Daily maximum 2-meter air temperature (°C) |
tmin |
Daily minimum 2-meter air temperature (°C) |
prcp |
Daily total precipitation (mm/day) |
srad |
Incident shortwave radiation flux density (W/m2) |
vp |
Water vapor pressure (Pa) |
swe |
Snow water equivalent (kg/m2) |
dayl |
Duration of the daylight period (seconds) |
Notes:
The Daymet calendar is based on a standard calendar year. All Daymet years, including leap years, have 365 days. Keep this in mind during post-processing of model results! For leap years, the Daymet database includes leap day (February 29) and values for December 31 are discarded from leap years to maintain a 365-day year.
DayMet’s incident shortwave radiation is the “daylit” radiation. To get the daily average radiation, one must multiply by daylit fraction, given by dayl / 86400.
%matplotlib inline
%load_ext autoreload
%autoreload 2
import matplotlib.pyplot as plt
import matplotlib as mpl
mpl.rcParams['figure.dpi'] = 150
import logging,yaml
import numpy as np
import rasterio
import fiona
import os
import datetime
import watershed_workflow
import watershed_workflow.ui
import watershed_workflow.sources.manager_daymet
import watershed_workflow.daymet
import watershed_workflow.io
watershed_workflow.ui.setup_logging(1,None)
watershed_workflow.config.set_data_directory('../../data')
watershed_name = 'CoalCreek'
watershed_shapefile = f'../../data/examples/{watershed_name}/sources/shapefile/CoalCreek.shp'
config_fname = f'../../data/examples/{watershed_name}/processed/config.yaml'
# Load the dictionary from the file
with open(config_fname, 'r') as file:
config = yaml.load(file, Loader=yaml.FullLoader)
# dates are in YYYY-MM-DD
# start and end of simulation -- one year of simulation that is in both the MODIS and DayMet dataset ranges
start_date = config['start_date']
end_date = config['end_date']
origin_date = config['origin_date']
logging.info(f"Daymet start date:{start_date} and end date:{end_date}. All times are relative to {origin_date}")
2025-07-12 18:38:31,939 - root - INFO: Daymet start date:2015-10-1 and end date:2017-10-1. All times are relative to 1980-1-1
import watershed#
crs, watershed = watershed_workflow.get_split_form_shapes(watershed_shapefile)
logging.info(f'crs: {crs}')
bounds = watershed.exterior().bounds
print(bounds)
print(bounds[2] - bounds[0], bounds[3] - bounds[1])
2025-07-12 18:38:32,023 - root - INFO:
2025-07-12 18:38:32,027 - root - INFO: Loading shapes
2025-07-12 18:38:32,030 - root - INFO: ------------------------------
2025-07-12 18:38:32,033 - root - INFO: Loading file: '../../data/examples/CoalCreek/sources/shapefile/CoalCreek.shp'
2025-07-12 18:38:32,194 - root - INFO: ... found 1 shapes
2025-07-12 18:38:32,197 - root - INFO: Converting to shapely
2025-07-12 18:38:32,208 - root - INFO: ... done
2025-07-12 18:38:32,220 - root - INFO: Removing holes on 1 polygons
2025-07-12 18:38:32,224 - root - INFO: -- removed interior
2025-07-12 18:38:32,230 - root - INFO: -- union
2025-07-12 18:38:32,233 - root - INFO: Parsing 1 components for holes
2025-07-12 18:38:32,236 - root - INFO: -- complete
2025-07-12 18:38:32,243 - root - INFO: crs: epsg:26913
(317251.2640131897, 4299711.408984916, 328473.7039815487, 4307062.45088187)
11222.439968359016 7351.041896954179
watershed.exterior()
Download#
returned raw data has dim(nband, ncol, nrow)
# setting vars = None to download all available variables
source = watershed_workflow.sources.manager_daymet.FileManagerDaymet()
data = source.get_data(bounds, crs, start = start_date, end = end_date, buffer=0.02)
2025-07-12 18:38:32,484 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,490 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_tmin_2015_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,493 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,498 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_tmin_2016_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,501 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,507 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_tmin_2017_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,510 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,521 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_tmax_2015_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,523 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,530 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_tmax_2016_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,533 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,538 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_tmax_2017_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,541 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,547 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_prcp_2015_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,550 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,556 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_prcp_2016_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,559 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,565 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_prcp_2017_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,568 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,572 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_srad_2015_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,575 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,580 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_srad_2016_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,582 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,587 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_srad_2017_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,590 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,595 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_vp_2015_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,598 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,626 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_vp_2016_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,630 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,641 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_vp_2017_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,644 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,650 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_dayl_2015_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,653 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,659 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_dayl_2016_38.9157x-107.1272_38.8072x-106.9560.nc
2025-07-12 18:38:32,661 - root - INFO: Collecting DayMet file to tile bounds: [-107.1272, 38.8072, -106.956, 38.9157]
2025-07-12 18:38:32,667 - root - INFO: Using existing: ../../data/meteorology/daymet/daymet_dayl_2017_38.9157x-107.1272_38.8072x-106.9560.nc
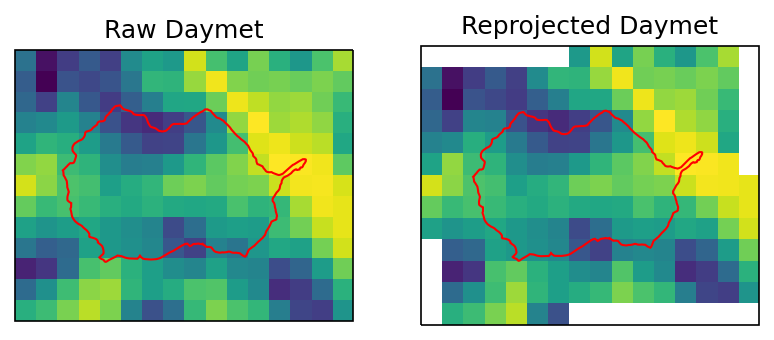
Reproject Daymet CRS#
Reproject daymet CRS to the same as the watershed. This is necessary if watershed meshes are using watershed CRS.
# this is a workaround, need to update the wrap.py to comment out 'dtype': src_profile['dtype']
data.profile.update({'dtype': data.data['prcp'].dtype})
data_warped = watershed_workflow.warp.dataset(data, dst_crs=crs)
plot Daymet#
ivar = 'tmax'
islice = 100
daymet_crs = watershed_workflow.crs.daymet_crs()
fig = plt.figure()
ax1 = watershed_workflow.plot.get_ax(daymet_crs, fig, 1, 2, 1)
ax2 = watershed_workflow.plot.get_ax(crs, fig, 1, 2, 2)
watershed_ext_daymet = watershed_workflow.warp.shply(watershed.exterior(),
crs, daymet_crs)
watershed_workflow.plot.raster(data[ivar].profile, data[ivar].data[islice,:,:], ax1)
watershed_workflow.plot.shply(watershed_ext_daymet, daymet_crs, ax=ax1, color='r')
watershed_workflow.plot.raster(data_warped[ivar].profile, data_warped[ivar].data[islice,:,:], ax2)
watershed_workflow.plot.hucs(watershed, crs, ax=ax2, color='r')
ax1.set_title("Raw Daymet")
ax2.set_title("Reprojected Daymet")
/opt/conda/envs/watershed_workflow/lib/python3.10/site-packages/pyproj/crs/crs.py:1282: UserWarning: You will likely lose important projection information when converting to a PROJ string from another format. See: https://proj.org/faq.html#what-is-the-best-format-for-describing-coordinate-reference-systems
proj = self._crs.to_proj4(version=version)
2025-07-12 18:38:36,244 - root - INFO: BOUNDS: (-591250.0, -367000.0, -576250.0, -355000.0)
2025-07-12 18:38:36,303 - root - INFO: BOUNDS: (314692.7009557779, 4296566.393670194, 331256.0668364619, 4310206.812630757)
Text(0.5, 1.0, 'Reprojected Daymet')
save daymet#
Write data to HDF5#
This will write daymet in a format that ATS can read. E.g., this will calculate mean air temperature, convert units, partition precipitation into rain and snow based on mean temp ( T_air=0 degC is used for the partitioning of rain and snow! This can be highly variable based on area. For more information on rain/snow partition, see this RainOrSnow website), and so on.
dout has dims of
(ntime, nrow, ncol)or(ntime, ny, nx)
met_data = watershed_workflow.daymet.convertToATS(data_warped)
config['daymet_filename'] = os.path.join('..', '..', 'data', 'examples', watershed_name, 'processed','watershed_daymet_raw.h5')
attrs = watershed_workflow.daymet.getAttributes(bounds, start_date, end_date)
attrs['name']="Daymet forcing reformatted for ats modeling"
watershed_workflow.io.write_dataset_to_hdf5(
filename=config['daymet_filename'],
attributes= attrs,
dataset=met_data,
time0 = origin_date
)
2025-07-12 18:38:36,953 - root - INFO: Converting to ATS met input
2025-07-12 18:38:36,996 - root - INFO: Writing HDF5 file: ../../data/examples/CoalCreek/processed/watershed_daymet_raw.h5
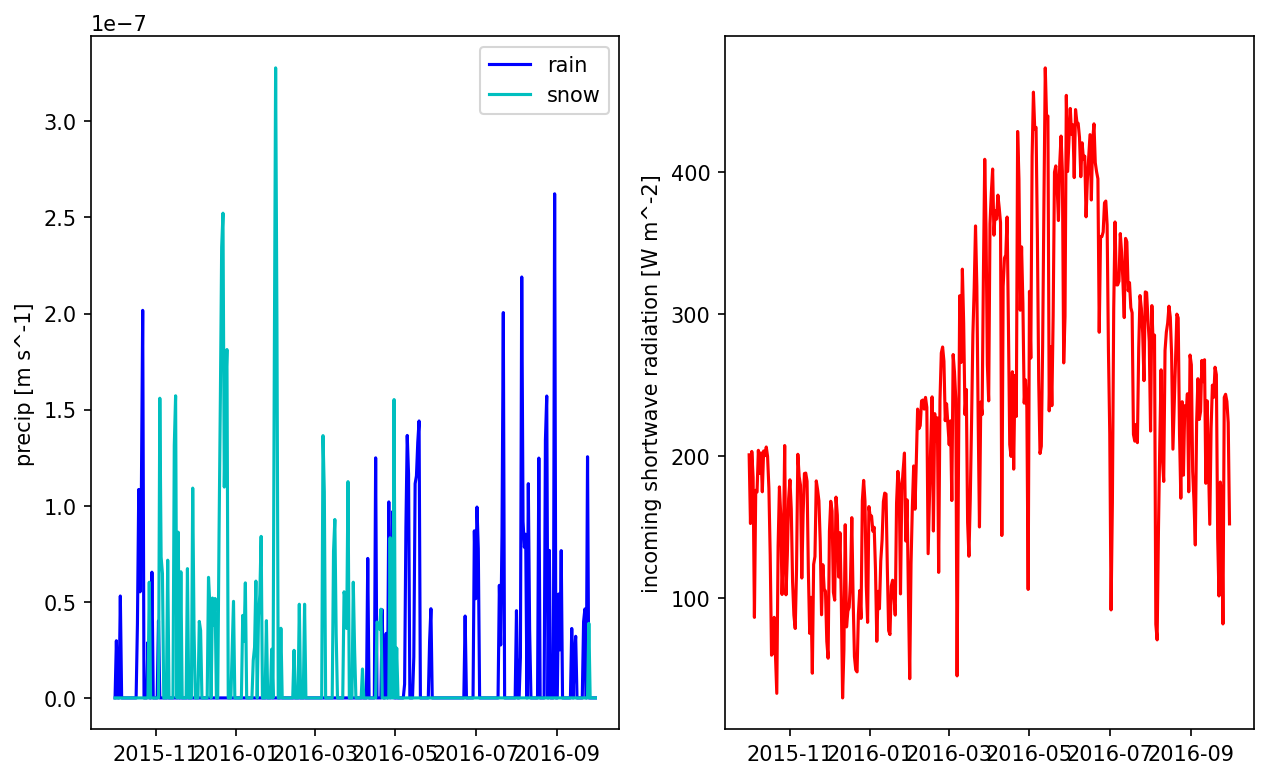
# plot one pixel as a function of time
fig = plt.figure(figsize=(10,6))
ax = fig.add_subplot(121)
times = np.array([datetime.datetime(*t.timetuple()[0:6]) for t in met_data.times])
prain = met_data['precipitation rain [m s^-1]'].data[:,5,5]
psnow = met_data['precipitation snow [m SWE s^-1]'].data[:,5,5]
ax.plot(times[0:365], prain[0:365], 'b', label='rain')
ax.plot(times[0:365], psnow[0:365], 'c', label='snow')
ax.set_ylabel('precip [m s^-1]')
ax.legend()
ax = fig.add_subplot(122)
qswin = met_data['incoming shortwave radiation [W m^-2]'].data[:,5,5]
ax.plot(times[0:365], qswin[0:365], 'r')
ax.set_ylabel('incoming shortwave radiation [W m^-2]')
plt.show()
Write typical Daymet#
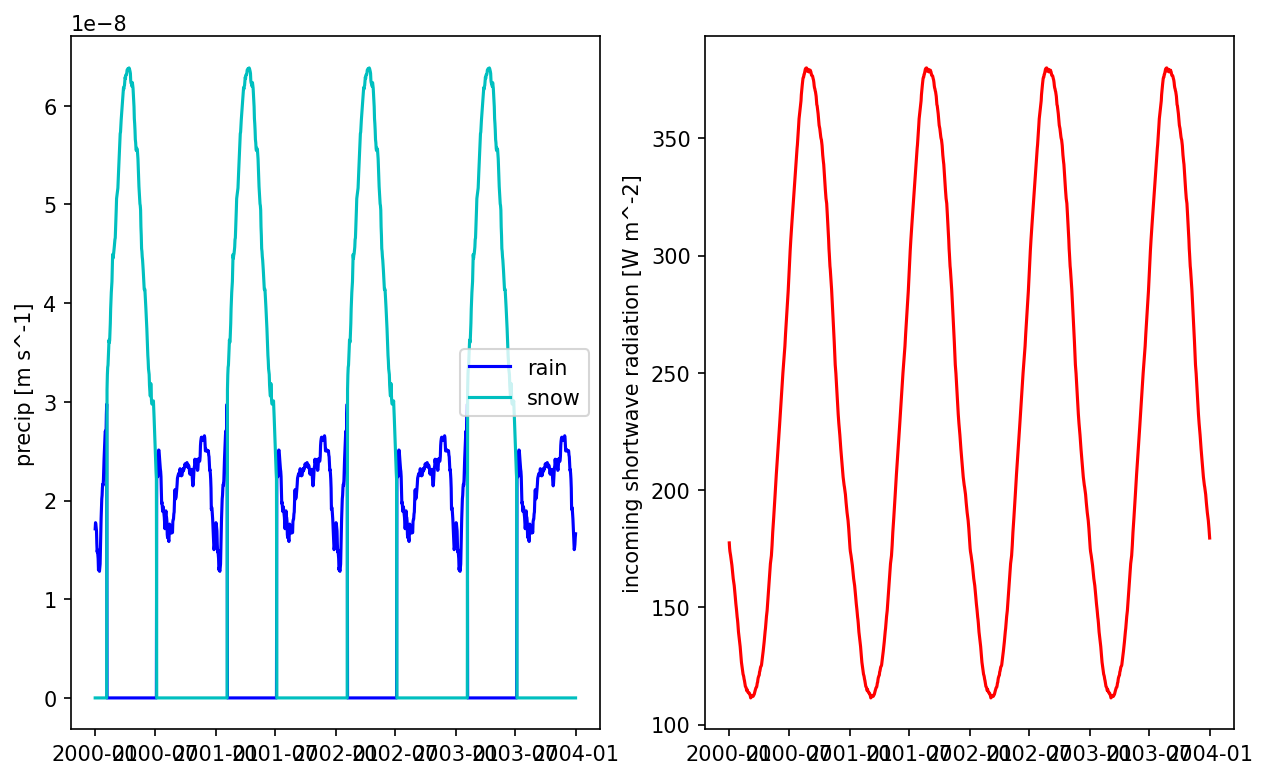
This will average days, smooths the forcing and generate a “typical year” data, then looped 10 years. The dataset will be used during cyclic spinup.
# compute the typical year of the _raw_ data
# note that we set interpolate to False, since met_data is already daily on a noleap calendar
nyears_cyclic_steadystate = config['nyears_cyclic_steadystate']
data_smooth = watershed_workflow.datasets.computeAverageYear(data_warped, output_nyears=nyears_cyclic_steadystate, smooth=True,
smooth_kwargs=dict(window_length=181, polyorder=2),
interpolate=False)
# convert that to ATS
met_data_smooth = watershed_workflow.daymet.convertToATS(data_smooth)
2025-07-12 18:38:48,066 - root - INFO: Converting to ATS met input
# plot the smoothed precip result
fig = plt.figure(figsize=(10,6))
ax = fig.add_subplot(121)
times = np.array([datetime.datetime(*t.timetuple()[0:6]) for t in met_data_smooth.times])
prain = met_data_smooth['precipitation rain [m s^-1]'].data[:,5,5]
psnow = met_data_smooth['precipitation snow [m SWE s^-1]'].data[:,5,5]
ax.plot(times, prain, 'b', label='rain')
ax.plot(times, psnow, 'c', label='snow')
ax.set_ylabel('precip [m s^-1]')
ax.legend()
ax = fig.add_subplot(122)
qswin = met_data_smooth['incoming shortwave radiation [W m^-2]'].data[:,5,5]
ax.plot(times, qswin, 'r')
ax.set_ylabel('incoming shortwave radiation [W m^-2]')
plt.show()
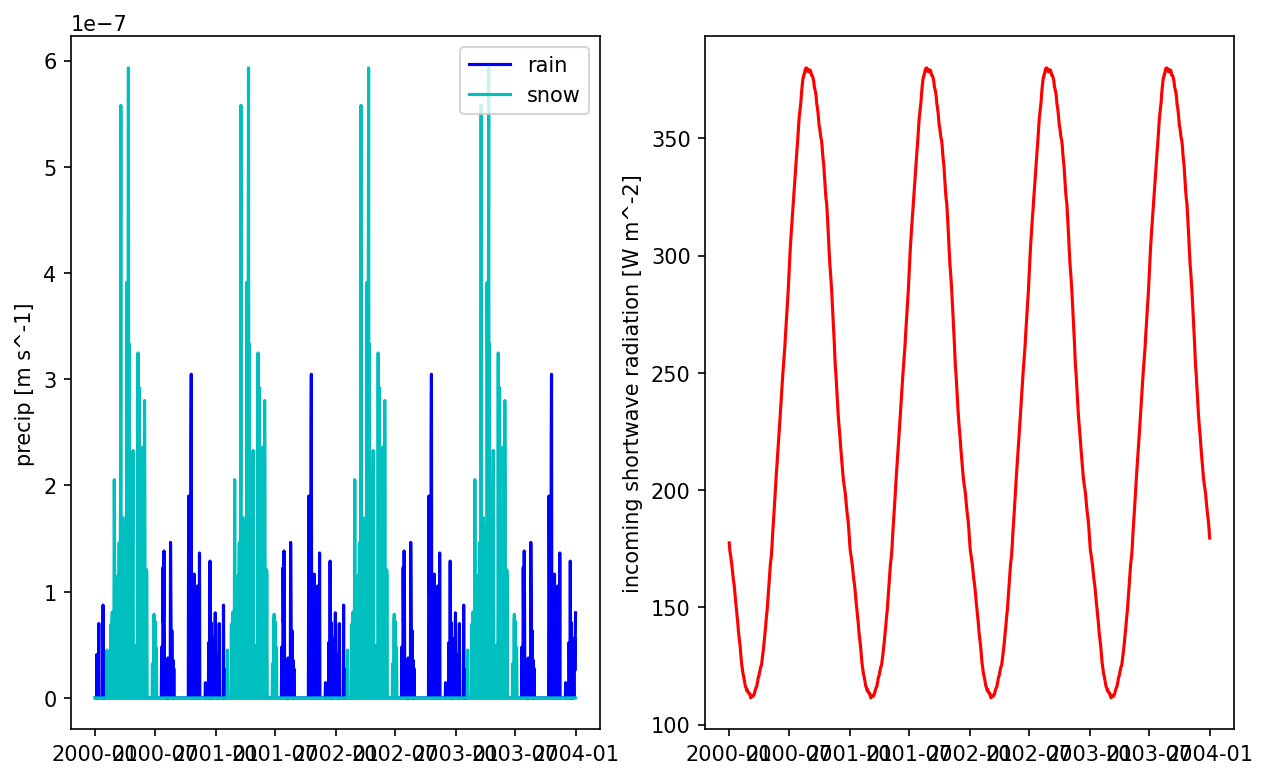
# often smoothing precip like that is a bad idea -- you now have every day misting with low intensity rain which can result in a ton of interception
# and canopy evaporation and no transpiration. Another approach is to just take the median total rainfall year and repeat that year multiple times.
precip_raw = data_warped['prcp'].data
shape_xy = precip_raw.shape[1:]
precip_raw = precip_raw.reshape((-1, 365,)+shape_xy)
annual_precip_raw = precip_raw.sum(axis=(1,2,3))
# note -- don't use np.median here... for even number of years it will not appear. Instead, sort and talk the halfway point
median_i = sorted(((i,v) for (i,v) in enumerate(annual_precip_raw)), key=lambda x : x[1])[len(annual_precip_raw)//2][0]
typical_precip_raw = precip_raw[median_i]
data_smooth['prcp'] = np.tile(typical_precip_raw, (nyears_cyclic_steadystate,1,1))
# np.tile(precip_raw[median_i], (2,1,1))
# data_smooth['prcp'] = typical_precip_raw
# convert that to ATS
met_data_smooth = watershed_workflow.daymet.convertToATS(data_smooth)
2025-07-12 18:38:50,226 - root - INFO: Converting to ATS met input
# plot the smoothed precip result
fig = plt.figure(figsize=(10,6))
ax = fig.add_subplot(121)
times = np.array([datetime.datetime(*t.timetuple()[0:6]) for t in met_data_smooth.times])
prain = met_data_smooth['precipitation rain [m s^-1]'].data[:,5,5]
psnow = met_data_smooth['precipitation snow [m SWE s^-1]'].data[:,5,5]
ax.plot(times, prain, 'b', label='rain')
ax.plot(times, psnow, 'c', label='snow')
ax.set_ylabel('precip [m s^-1]')
ax.legend()
ax = fig.add_subplot(122)
qswin = met_data_smooth['incoming shortwave radiation [W m^-2]'].data[:,5,5]
ax.plot(times, qswin, 'r')
ax.set_ylabel('incoming shortwave radiation [W m^-2]')
plt.show()
# write to disk
config['daymet_typical_filename'] = os.path.join('..', '..', 'data', 'examples', watershed_name, 'processed','watershed_daymet_typical.h5')
attrs = watershed_workflow.daymet.getAttributes(bounds, start_date, end_date)
# attrs['name']="Daymet forcing reformatted for ats modeling"
attrs['name']=f"{nyears_cyclic_steadystate}-yrs typical Daymet forcing generated using data from {start_date} to {end_date}"
watershed_workflow.io.write_dataset_to_hdf5(
filename=config['daymet_typical_filename'],
dataset=met_data_smooth,
attributes= attrs,
time0 = None) # use the first date as the time0, relative time to be consistent with start time in xml file
2025-07-12 18:38:52,351 - root - INFO: Writing HDF5 file: ../../data/examples/CoalCreek/processed/watershed_daymet_typical.h5
ivar = 'tmax'
islice = 100
# daymet_crs = watershed_workflow.crs.daymet_crs()
fig = plt.figure()
ax1 = watershed_workflow.plot.get_ax(crs, fig, 1, 2, 1)
ax2 = watershed_workflow.plot.get_ax(crs, fig, 1, 2, 2)
# watershed_ext_daymet = watershed_workflow.warp.shply(watershed.exterior(),
# crs, daymet_crs)
# watershed_workflow.plot.raster(data_warped[ivar].profile, data_warped[ivar].data[islice,:,:], ax1)
# watershed_workflow.plot.shply(watershed_ext_daymet, daymet_crs, ax=ax1, color='r')
watershed_workflow.plot.raster(data_warped[ivar].profile, data_warped[ivar].data[islice,:,:], ax1)
watershed_workflow.plot.hucs(watershed, crs, ax=ax1, color='r')
watershed_workflow.plot.raster(data_smooth[ivar].profile, data_smooth[ivar].data[islice,:,:], ax2)
watershed_workflow.plot.hucs(watershed, crs, ax=ax2, color='r')
ax1.set_title("Reprojected Daymet")
ax2.set_title("Smoothed Daymet")
2025-07-12 18:39:07,386 - root - INFO: BOUNDS: (314692.7009557779, 4296566.393670194, 331256.0668364619, 4310206.812630757)
2025-07-12 18:39:07,413 - root - INFO: BOUNDS: (314692.7009557779, 4296566.393670194, 331256.0668364619, 4310206.812630757)
Text(0.5, 1.0, 'Smoothed Daymet')
Get mean precipitation for spinup
# calculate the basin-averaged, annual-averaged precip rate
precip_total = met_data.data['precipitation rain [m s^-1]'] + met_data.data['precipitation snow [m SWE s^-1]']
mean_precip = precip_total.mean()
config['mean_precip [m s^-1]'] = float(mean_precip)
logging.info(f'Mean annual precip rate [m s^-1] = {mean_precip}')
2025-07-12 18:39:07,973 - root - INFO: Mean annual precip rate [m s^-1] = 2.4889101869462758e-08
with open(config_fname, 'w') as file:
yaml.dump(config, file)